Quick Tour

Please watch the following video for a quick demo on how to search for symbiotic organisms in SymGenDB and/or head over to the step by step tutorial beneath the demo video for more information.
NOTE: These videos were made with a previous version of SymGenDB. Don't be surprised if you get more results!
Step by step quick tour:
1. Chose which partner of the symbiosis you want to search for. It can either be the symbionts of your host of interest, or the other way around.

2. Write the popular-name/specie/class/genus/(etc) of your interest according to the taxonomical level you are interested in, in the white search box.
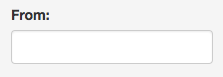
3. Select the host(s) taxonomy level you are interested in (the default ‘all ranks’ will search in every taxonomic level and as a result give you the level where the search is found).
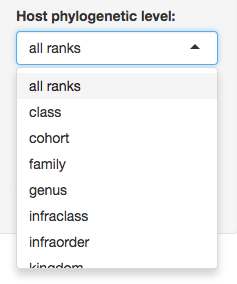
4. Select the symbiont(s) taxonomy level you are interested in (the default ‘all ranks’ will search in every taxonomic level and as a result give you the level where the search is found).
5. Typically, in just a few seconds you will be able to see the results of your query. The results include an abundance graph of the organisms in the symbiotic relationship(s), and a table listing every specie, order, class, phylum, or any taxonomy level selected.
[Example. In our laboratory at the University of Valencia, we love insects. Here is the result of searching for the symbionts of ‘insects' with the default 'all ranks' host level, and the ‘species' level selected for symbionts.]
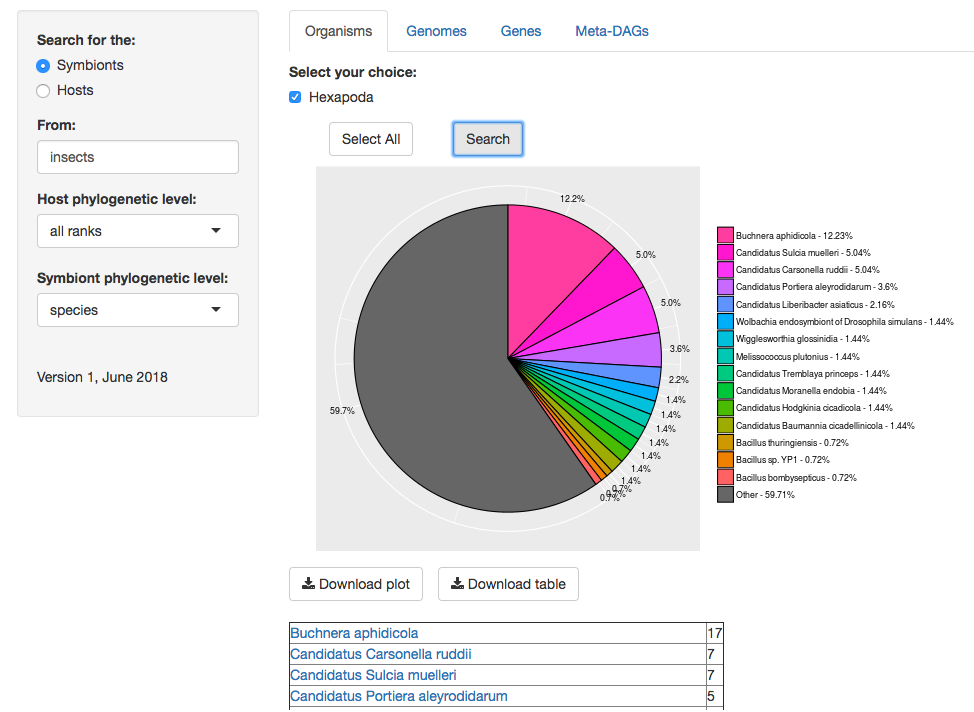
(the table of organisms goes on until every specie found is listed. We only show the first part of the list).
6. Download the plot and/or table resulting of your search if needed.
7. Be aware that the resulting list of organisms (in the example above, the species involved in symbiotic relationships with insects) are links. These links direct to each species' NCBI Taxonomy Browser.

Please watch the following video for a quick example on how to search for the genomes of symbiotic organisms in SymGenDB and/or head over to the step by step tutorial after the demo video.
Step by step quick tour:
1. Write in the white search box the popular-name/specie/class/genus/(etc) of your genome(s) of interest according to the taxonomical level you are interested in (the default ‘all ranks’ will search in every taxonomic level). You can search for a specific symbiont, or a specific host. If you search for a host, you will have another menu listing all its symbiontic genomes available, where the genomes of interest can be selected.
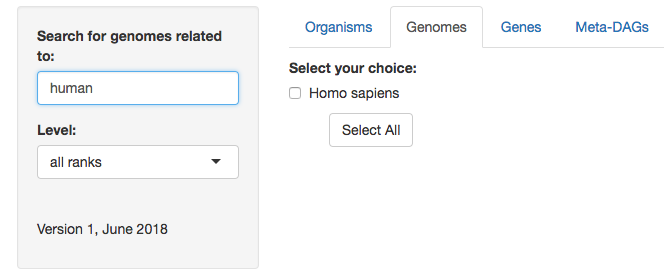
2. Select the symbiont(s) genome(s) you are interested in by selecting the box(es) available. In this example, first for the only match of human (Homo sapiens) and then for the genomes you want to explore. You can also 'select all'.
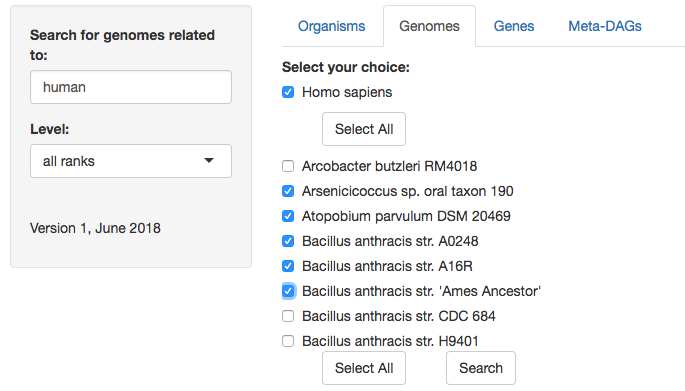
3. Typically, in just a few seconds you will be able to see the results of your query. The table displayed for each symbiont consist of the name of the host it belongs to, and the metrics of each genome selected, including plasmids and all chromosomes in cases where more than one chromosome is present.
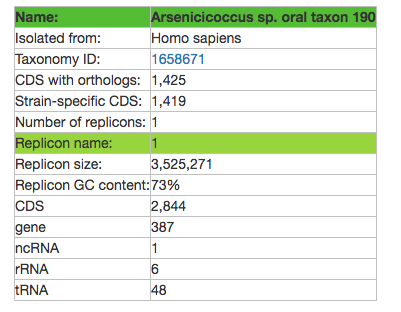
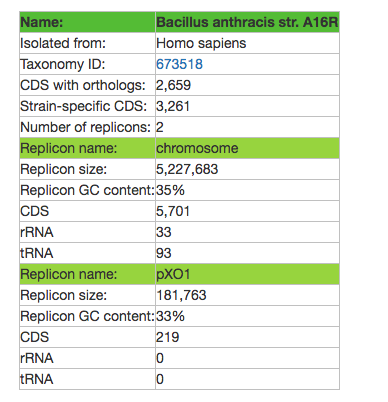
NOTE: You also get a counter telling you how many genomes are symbionts of the host you searched for, and how many you selected (in this case, for human).
4. Download the table(s), the genomic FASTA file(s), the GIFF file(s) and/or the ortholog table of the organisms resulting of your search if needed.  Note: With these results, an orthology table is created between the resulting genomes of each search. This orthology table includes different fields of information for each ortholog found, including the gene name, function and E.C. number. Then, each organism of the search is displayed as a column (abbreviated with their KEGG organism id as the header of the column), with a list of each ortholog shared with at least one other organism in the subset selected.
Note: With these results, an orthology table is created between the resulting genomes of each search. This orthology table includes different fields of information for each ortholog found, including the gene name, function and E.C. number. Then, each organism of the search is displayed as a column (abbreviated with their KEGG organism id as the header of the column), with a list of each ortholog shared with at least one other organism in the subset selected.

Please watch the following video for a quick example on how to search for the genes of symbiotic organisms in SymGenDB and/or head over to the step by step tutorial after the demo video.
Step by step quick tour:
1. Write the name of the gene(s) you are interested in. Use commas to separate between genes of interest if more than one is searched for.
2. Write the popular-name/specie/class/genus/(etc) of your genome(s) of interest according to the taxonomical level you are interested in, in the white search box (the default ‘all ranks’ will search in every taxonomic level).
3. You can search for a specific symbiont, or a specific host. If you search for a host, you will have another menu listing all its symbionts’ genomes available, where the genomes of interest can be selected.
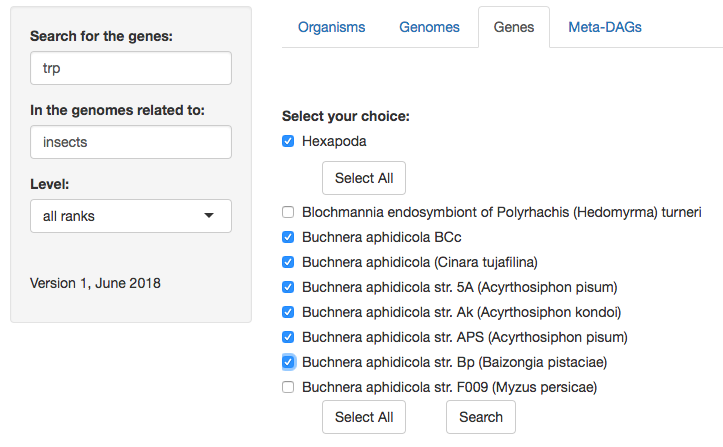
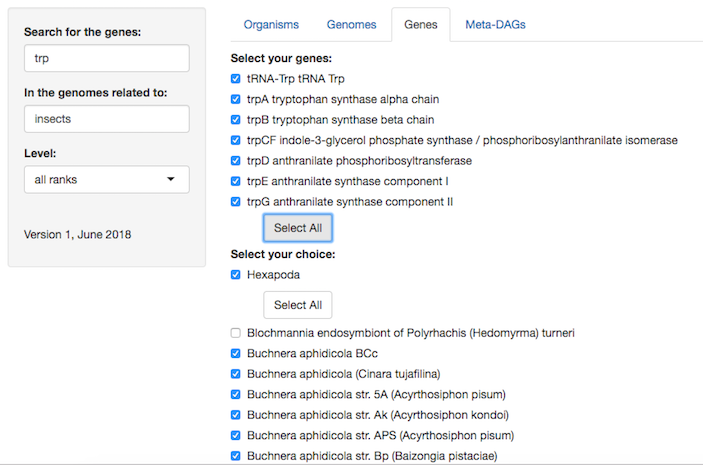
4. Then, search for the gene(s) you are interested in or 'select all'.
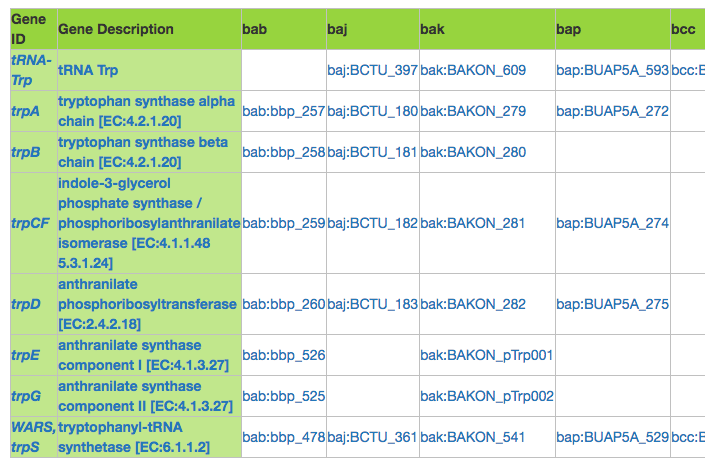
5. The resulting table will list the orthologous genes found between the genomes selected. We have used the KEGG nomenclature to denote the organisms' names on the table for spacing purposes. All the results are llinks. The first two columns will lead you to the KEGG Orthology web page of each result, and the specific ids for each organism will lead you to the specific gene web page. If you have a large list of organisms, you should scroll to the right to find all the data resulting of your search.
6. The resulting table of orthologs, as well as their peptide sequences, are downloadable.
Please watch the following video for a quick example on how to search for the Meta-DAGs of symbiotic organisms in SymGenDB and/or head over to the step by step tutorial after the demo video.
Step by step quick tour:
For this tab, we have calculated the 'reduced metabolic networks' (Meta-DAGs) of all the organisms included in SymGenDB based on the methodology by Ricardo Alberich, José A. Castro, Mercè Llabrés and Pere Palmer-Rodríguez from the University of the Balearic Islands, featured in their paper: Metabolomics analysis: Finding out metabolic building blocks.
We strongly encourage users to read the aforementioned paper to undersand all the details of this technique, but the gist of it is that this methodology aims to reduce the size of metabolic networks by calculating 'Metabolic Building Blocks' (MBBs) based on the ciclyc and acyclic properties of the pathways included in an organism's metabolic network, for easier understanding, analysis and interpretations. It is also a robust tool for the comparisson of Meta-DAGs between 2 or more organisms. For this reason, we have included two types of analysis. First, the calculation of each organism's Meta-DAG, and second, the calculation of the combined Meta-DAGs of strains included in the taxonomic level: genus. For this case, the output includes two different analysis, the common metabolism of all the strains included in the genus/genera of interest ('core Meta-DAGs') and the joint metabolism of these strains ('pan Meta-DAGs').
The output of this searches is a link that conducts the user to an interactive graph of the Meta-DAG of the organism searched for, and two other links for every hit, one to the core Meta-DAG and one to the pan Meta-DAG of the genus to which the searched bacteria belongs to, as explained before. This dynamic graph is easy to understand, users can zoom in and out of the Meta-DAG to get either the overview or the details of the graph, as well as click on any of its MMBs, to get the details of the reaction(s) that make up that MBB. Every reaction in the MBBs is linked to it's KEGG reaction web page, and also, a list of other pathways where the reaction also participates is presented.
1. Write the popular-name/specie/class/genus/(etc) of your interest according to the taxonomical level you are interested in, in the white search box.
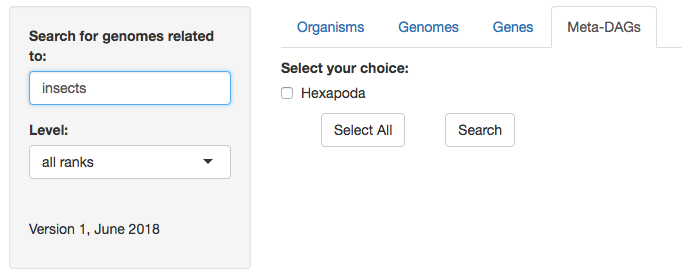
2. Select the organism(s) you are interested in. For this example we will search for the symbionts of insects again (that's how much we love them!).
3. As a result, you will get a list of all the symbionts related to the organism(s) you searched for:
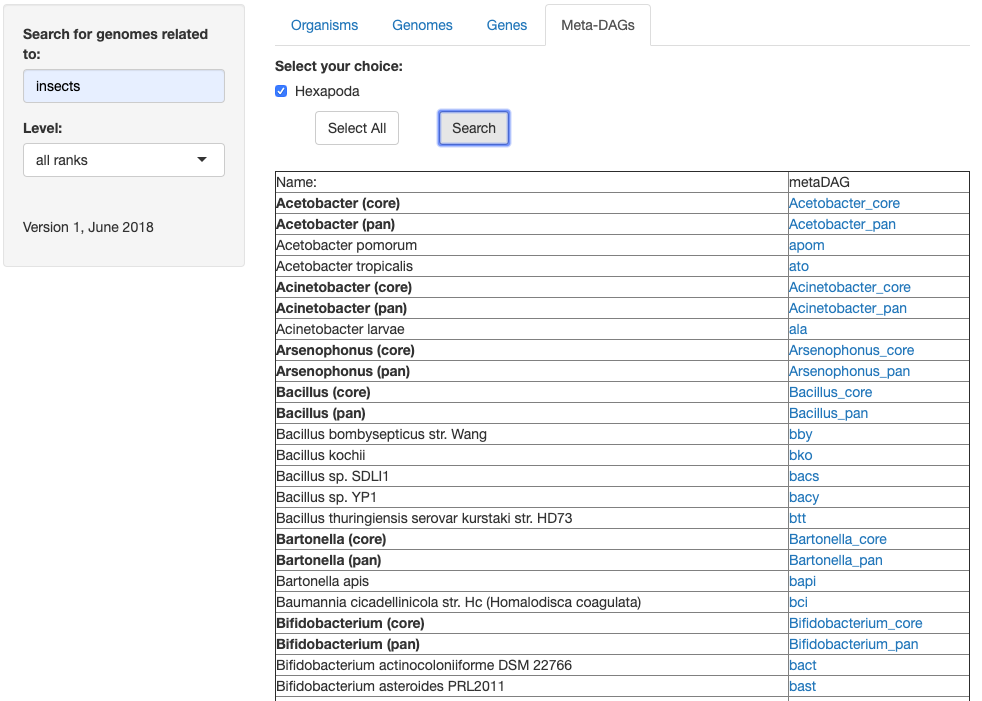
4. This list consists of the symbiont strains included in the symbiotic relationship, as well as the common (core) and joint (pan) Meta-DAGs of ALL the strains included in the genus (IMPORTANT NOTE: the genus Meta-DAGs include ALL the strains in SymGenDB that belong to the genus, not only the ones that are symbionts to the organisms you originally searched for).
In BOLD you will find the genera with both options in different rows (core and pan).

When you click on the image file of any of the hits you get, you are able to see a preview of the Meta-DAG, and you can instantly explore it. You can zoom in and out, you can click on any MBB and see the reation(s) that comprise it, and you can click on any reaction to go to its web page in KEGG. There are two buttons at the top of the window where you can open the same Meta-DAG in a new tab of your internet explorer, and download the image. This example is the core metabolism of all the bacillus strains included in SymGenDB:
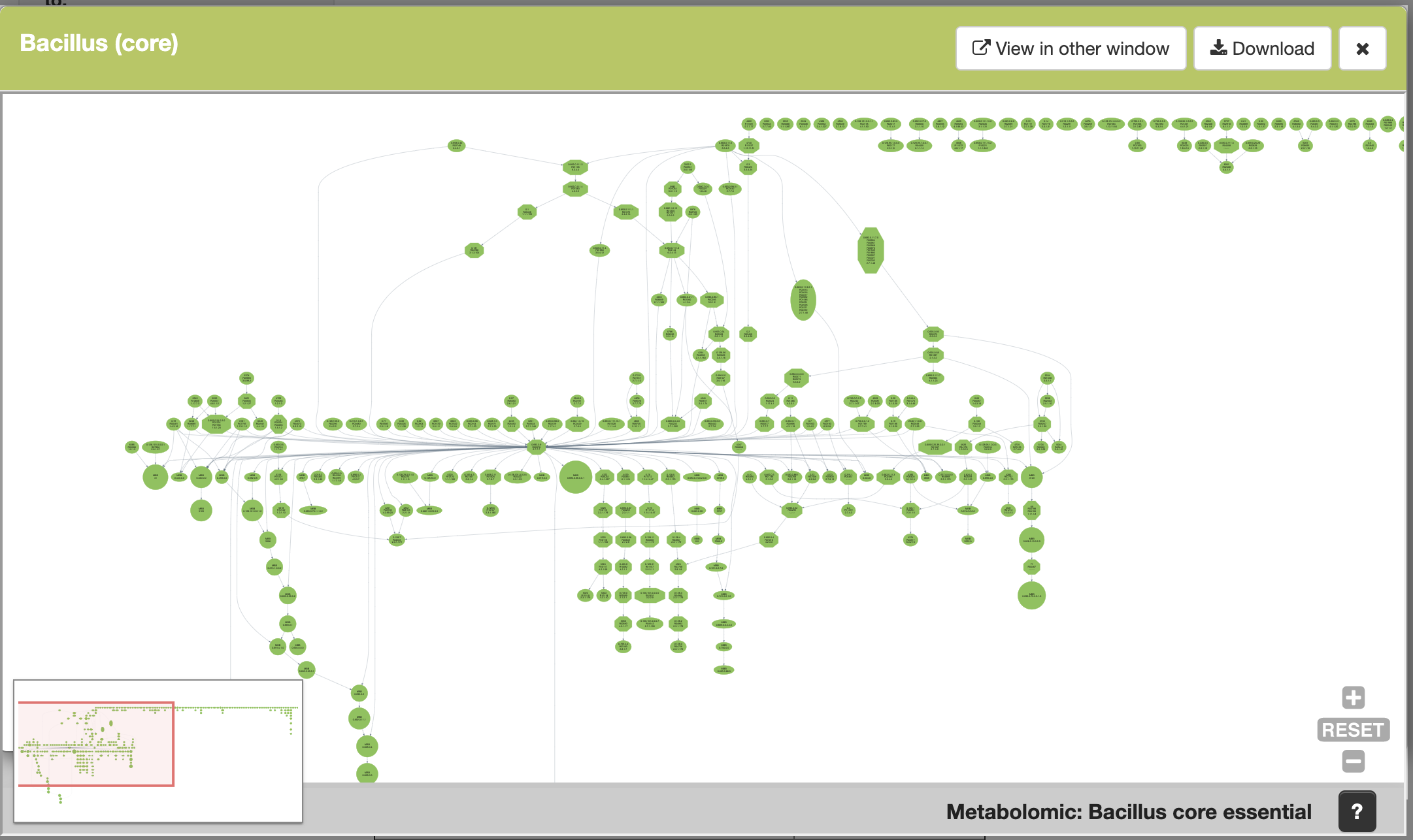
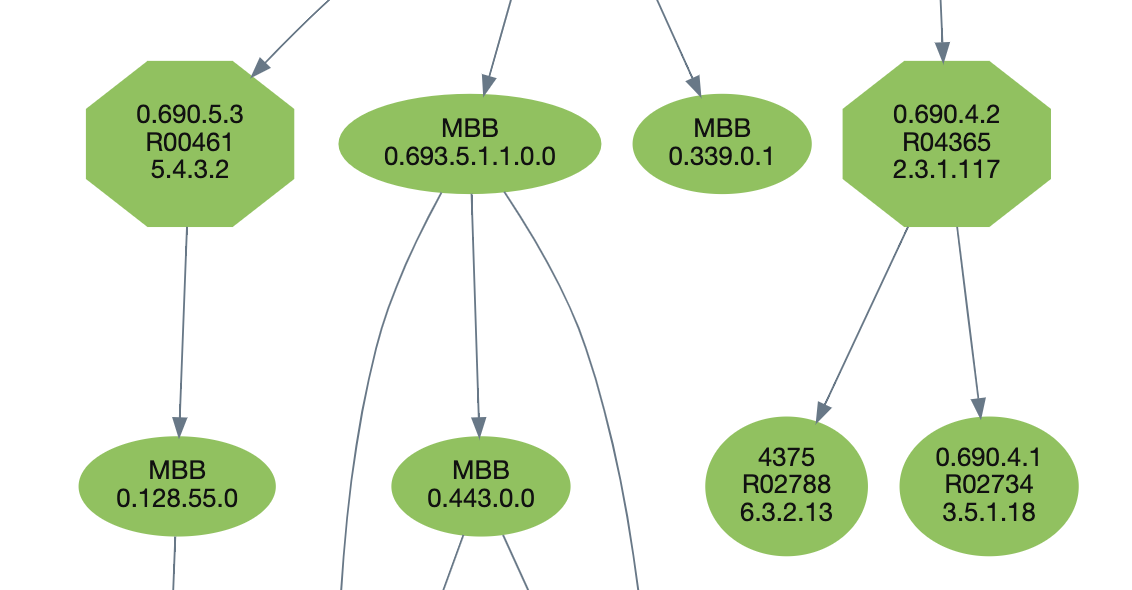
5. Regarding shapes, there are two types in Meta-DAGs: circles which include one or more reactions (you can easily get an idea by the size of the circle), and octagons, which denote what we call essential reactions, those that can break up the network's connectivity.
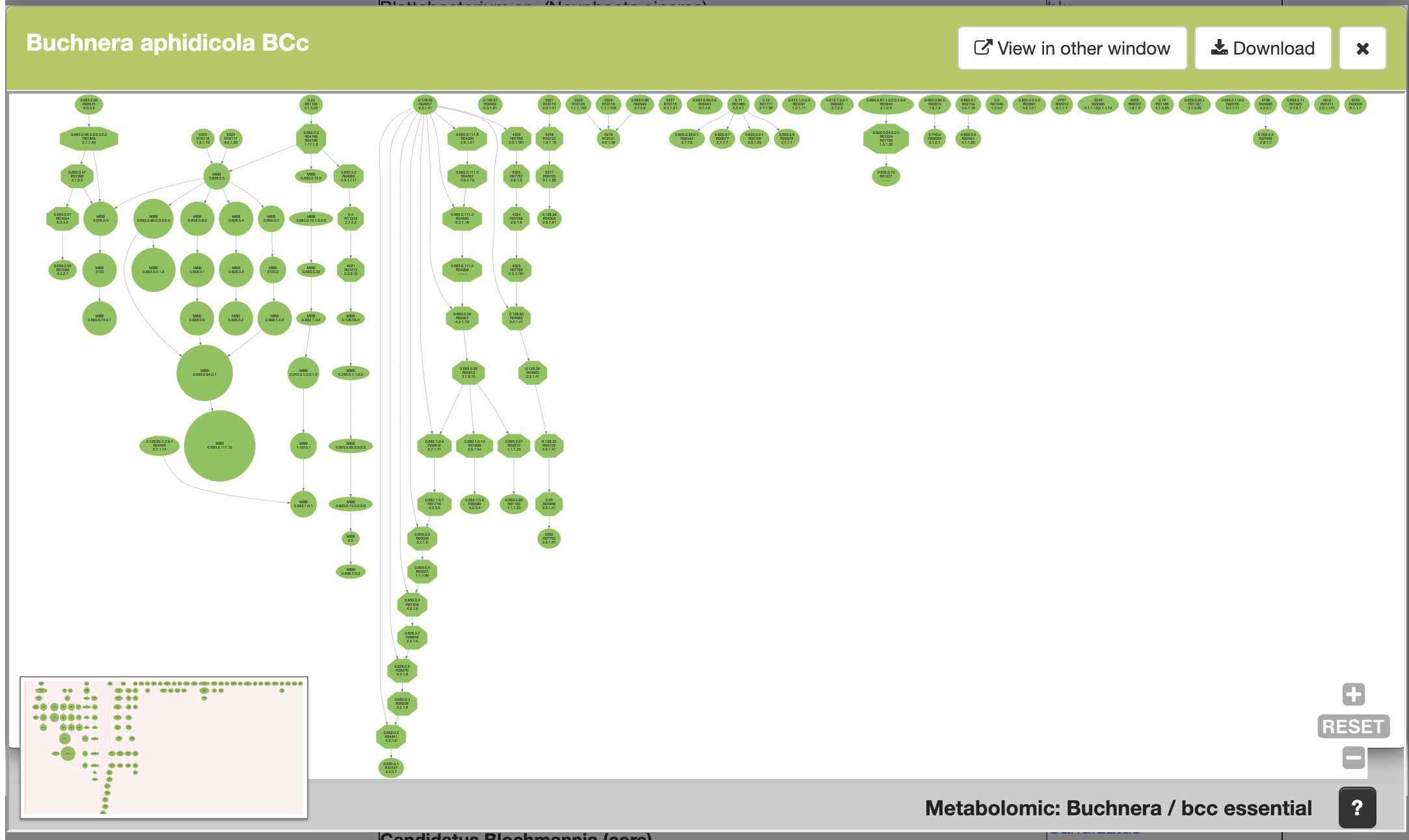
This is another example, the Meta-DAG of Buchnera aohidicola BCc.
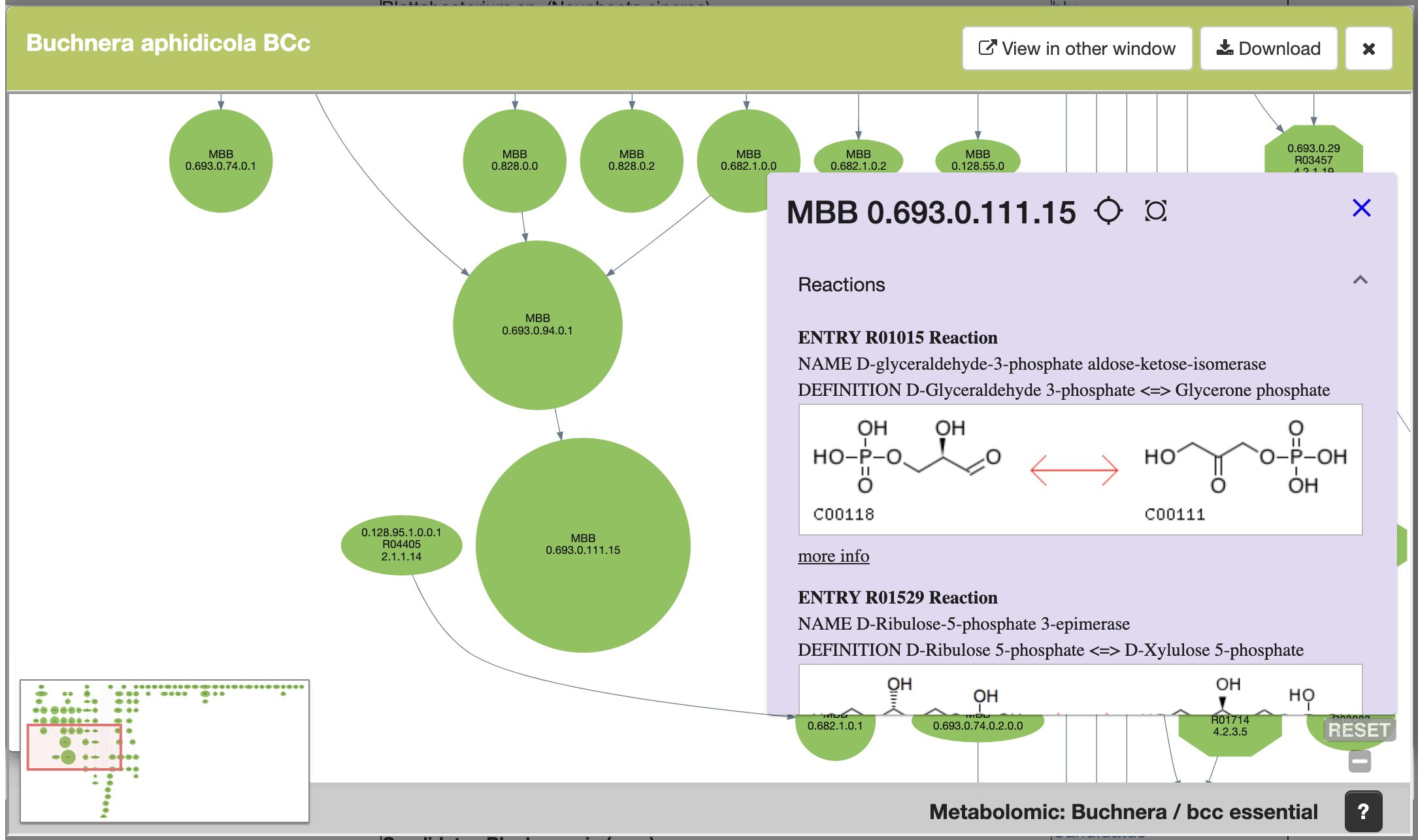
Once you click on a MBB, you get access to the reaction(s) that makes up that MBB and to the(ir) KEGG ids, and their KEGG reaction web page by clicking on "more info", as well as a list of other graphs where this reaction participates.
A new and exciting feature we have included in our database, is the possibility of looking at these reacions in context, meaning, highlighting the reactions of interest in the complete metabolism map by KEGG.
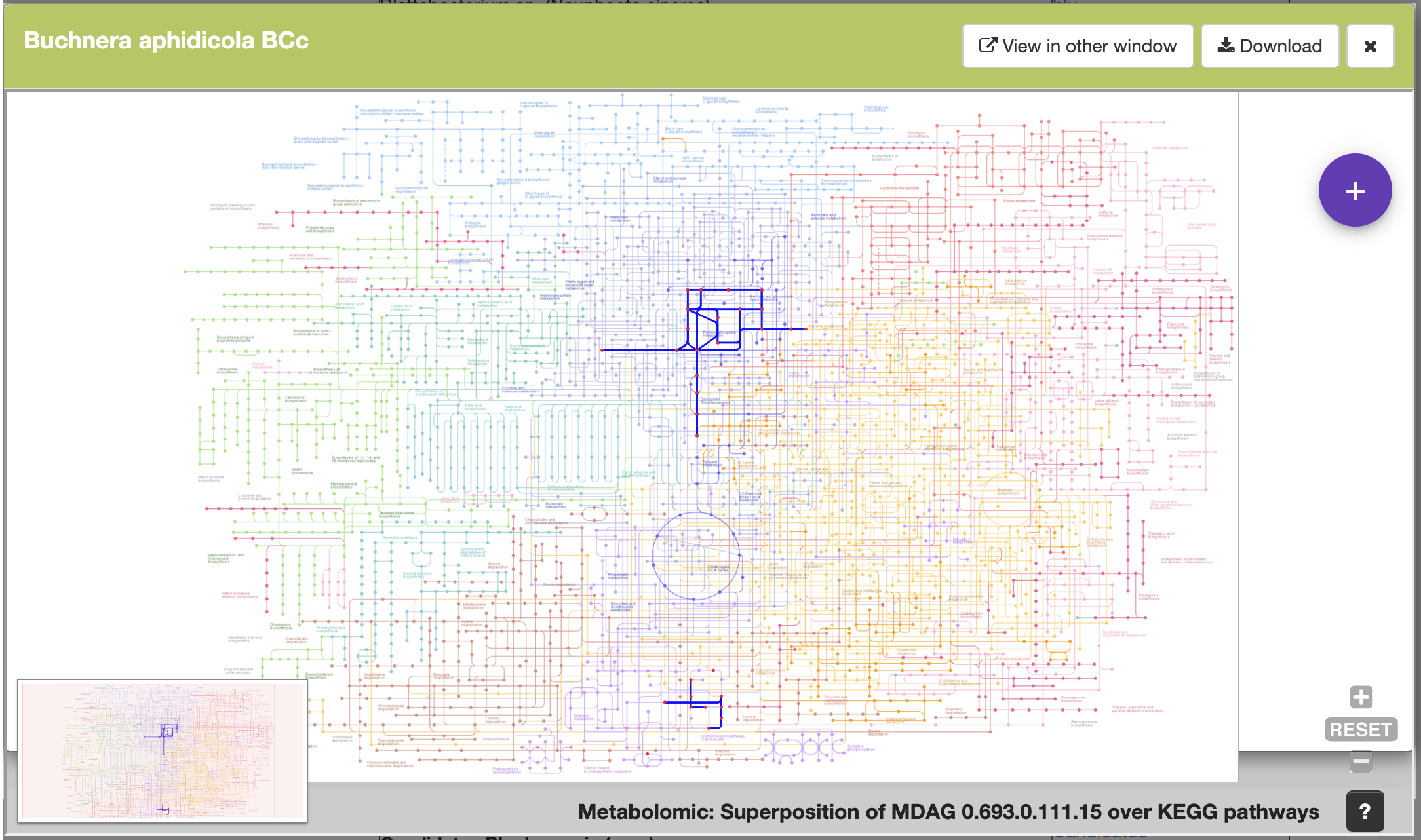
Also, users are able to get to the reaction's web page by clicking on the "more info" link. Please refer to the quicktour video for more info.