Symbiotic relationships occur naturally throughout the tree of life, either in a commensal, mutualistic or pathogenic manner. The genomes of multiple organisms involved in symbiosis are quickly being sequenced and becoming available, especially those in the microbial world. There are currently numerous databases that offer information of specific organisms or models, but none offer a global understanding on symbiosis and their community complementation and capabilities.
Symbiotic relationships occur naturally throughout the tree of life, either in a commensal, mutualistic or pathogenic manner. The genomes of multiple organisms involved in symbiosis are quickly being sequenced and becoming available, especially those in the microbial world. There are currently numerous databases that offer information of specific organisms or models, but none offer a global understanding on symbiosis and their community complementation and capabilities.
SymGenDB consists of a community database resource for laboratories which aim to research and gather information of the genetics and the genomic systems involved in symbiosis, regardless of it’s biotic interaction as commensal, mutualist or pathogen.
SymGenDB maintains a catalogue of sequenced/finished genomes of symbionts from organisms throughout the tree of life, as well as their genomic features. The catalogue was generated with computational tools, custom R scripts, and manual integration of information available in public literature. As a highly curated and comprehensive systems database, SymGenDB provides web access to the catalogue of sequenced and annotated symbiotic genomes of symbiotic organisms, their features including genomic sequences and metrics, as well as links to central databases such as KEGG and NCBI.
Because
more data is being generated and included in mayor databases,
SymGenDB will be updated and capable
of providing information on
these symbiotic organisms, as to become an important, useful and
timesaving tool.
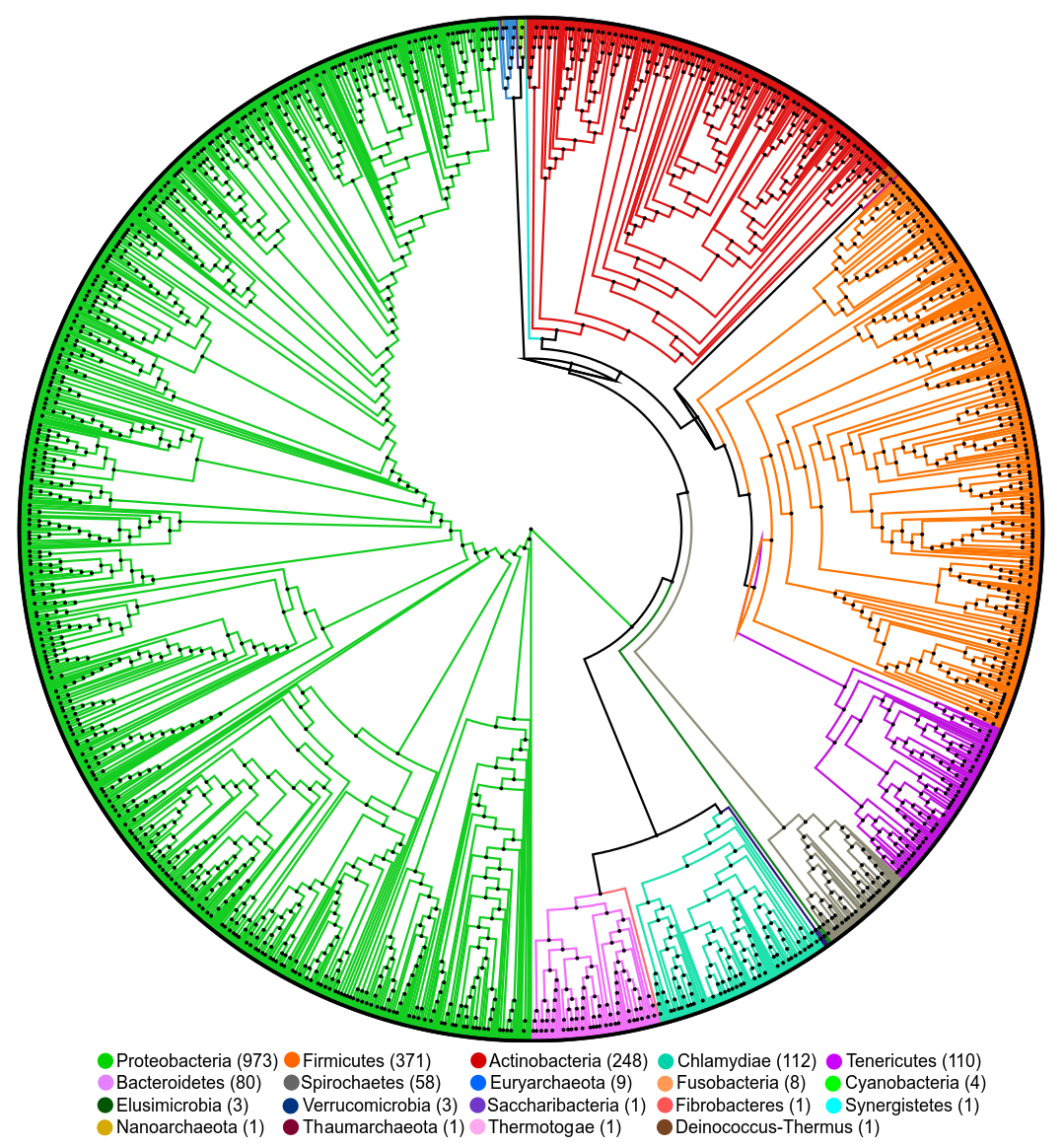
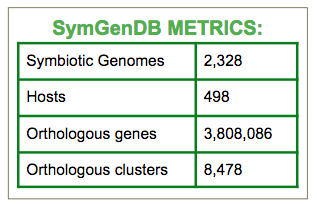
We currently have 2328 symbiotic genomes included in the database. An overview of the phyla comprising this list is
shown in the next phylogenetic tree (Constructed with the software PhyloPhlan).